

文獻(xiàn)信息:
Fierer Noah*. Embracing the unknown: disentangling the complexities of the soil microbiome[J]. Nature Reviews Microbiology, 2017, 15(10): 579-590. DOI:10.1038/nrmicro.2017.87
Nature Reviews Microbiology最新(2020年)影響因子:60.633
Key Points:
-
Soils can contain large amounts of microbial biomass, including fungi, protists, viruses, bacteria and archaea. Most of these taxa currently remain undescribed, and have physiological and ecological attributes that are unknown.
-
Soil microbial communities are highly diverse, in part because soil environmental conditions are so heterogeneous. In a single soil there is a wide range of distinct microbial habitats that contain unique microbial assemblages.
-
Spatial variability in the structure of soil microbial communities is typically larger than the temporal variability. The composition of soil bacterial communities and the abundances of specific taxa are often predictable from soil and site characteristics, including soil pH, climate and organic carbon availability.
-
Plants can clearly have important direct or indirect effects on soil microbial communities and vice versa. However, the effects of plant species on microbial taxa are often difficult to predict a priori owing, in part, to plant associations with soil microorganisms being highly context-dependent.
-
Linking specific soil microbial processes to specific microbial taxa remains difficult. One way to tackle this problem is to use genomic data to group microbial taxa according to shared similar life-history strategies and functional attributes.
-
Given recent methodological and conceptual advances, the field is poised to rapidly advance our understanding of the soil microbiome. Promising future research directions include cultivation-based analyses of soil microbial taxa, studies of soil viruses and investigations into the importance of horizontal gene transfer in shaping the soil microbiome.
摘 要:土壤微生物是自然生態(tài)系統(tǒng)和人為管理生態(tài)系統(tǒng)的關(guān)鍵組成部分。1 g土壤中可以涵蓋數(shù)千個(gè)微生物類群,包括病毒和所有三個(gè)生命域的成員。目前在標(biāo)記基因、基因組和宏基因組分析方面的最新進(jìn)展極大地?cái)U(kuò)展了我們對(duì)土壤微生物群落特征的研究,并確定了在空間和時(shí)間上形成土壤微生物群落的因素。然而,雖然大多數(shù)土壤微生物還沒有被精準(zhǔn)描述出來,但我們?nèi)钥梢愿鶕?jù)土壤微生物生態(tài)策略對(duì)其進(jìn)行分類。這是一種利用基因組信息預(yù)測(cè)單個(gè)微生物類群功能屬性的有效方法。該領(lǐng)域現(xiàn)在正準(zhǔn)備確定我們如何控制和管理土壤微生物群落,以提高土壤肥力、作物產(chǎn)量以及我們對(duì)陸地生態(tài)系統(tǒng)將如何響應(yīng)環(huán)境變化的理解。
Abstract: Soil microorganisms are clearly a key component of both natural and managed ecosystems. Despite the challenges of surviving in soil, a gram of soil can contain thousands of individual microbial taxa, including viruses and members of all three domains of life. Recent advances in marker gene, genomic and metagenomic analyses have greatly expanded our ability to characterize the soil microbiome and identify the factors that shape soil microbial communities across space and time. However, although most soil microorganisms remain undescribed, we can begin to categorize soil microorganisms on the basis of their ecological strategies. This is an approach that should prove fruitful for leveraging genomic information to predict the functional attributes of individual taxa. The field is now poised to identify how we can manipulate and manage the soil microbiome to increase soil fertility, improve crop production and improve our understanding of how terrestrial ecosystems will respond to environmental change.
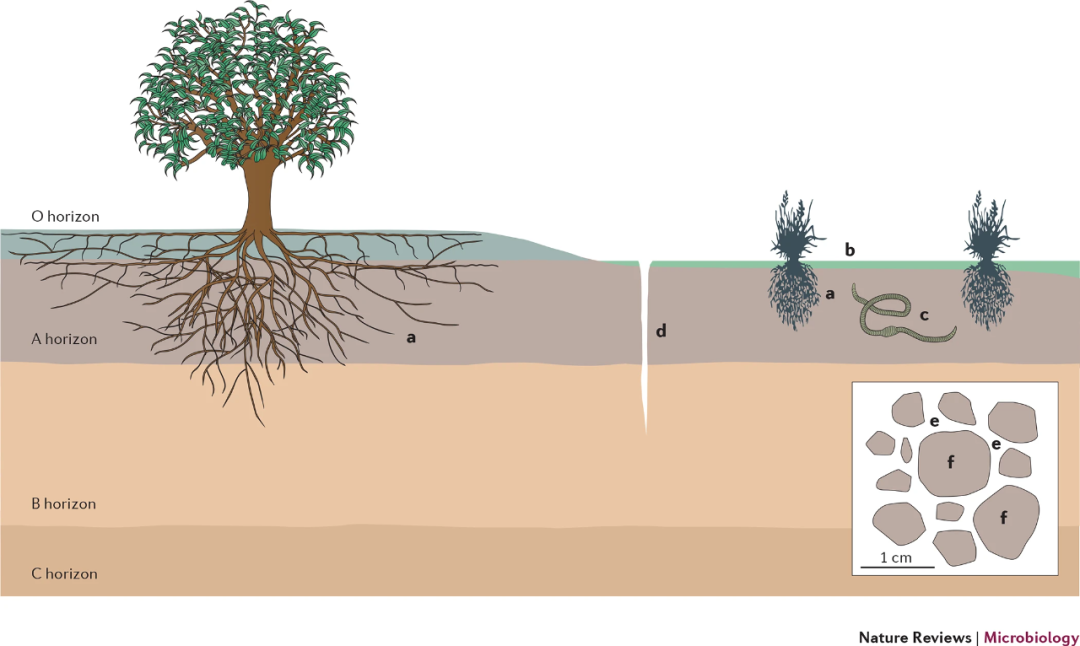
圖1 土壤的宏觀環(huán)境和微觀環(huán)境
Figure 1 The macroenvironments and microenvironments of soil. Soil is not a single environment; instead, soil encompasses a broad range of different microbial habitats. These include the rhizosphere (soil in close proximity to plant roots; part a), surface layers that are exposed to light (part b; the photic zone), soil associated with earthworm burrows (the drilosphere; part c), and soil found in preferential water flow paths, including cracks in the soil (part d). Moreover, there are microenvironments associated with soil aggregates; the conditions found on aggregate surfaces or on the water films between aggregates (part e) are distinct from the conditions found inside aggregates (part f). Finally, there are marked shifts in microbial communities and abiotic conditions with soil depth. Although most studies have focused exclusively on the microorganisms found in surface soil horizons (layers), communities found in the litter layer (or O-horizon) are often distinct from those found in underlying mineral soil horizons (A and B horizons) and deeper saprolite (C horizons). Key soil properties — such as pH, organic carbon concentration, salinity, texture and available nitrogen concentration — can vary substantially across these distinct soil environments.
圖2 土壤中發(fā)現(xiàn)的全球微生物生物量
Figure 2 Global microbial biomass found in soil. An estimation of how total soil microbial biomass (the sum of all microbial groups: bacteria, fungi, archaea, protists and viruses) varies across the globe (part a), and estimates of the contributions of the major microbial groups to this total soil microbial biomass pool (part b). The estimates in part b were compiled from information provided in Refs 104,124,125,126,127,128,129,130. These estimates are approximations; biomass can vary substantially across soils, and the biomass of protists and viruses is highly uncertain. The map in part a is adapted with permission from Ref. 2, Wiley.
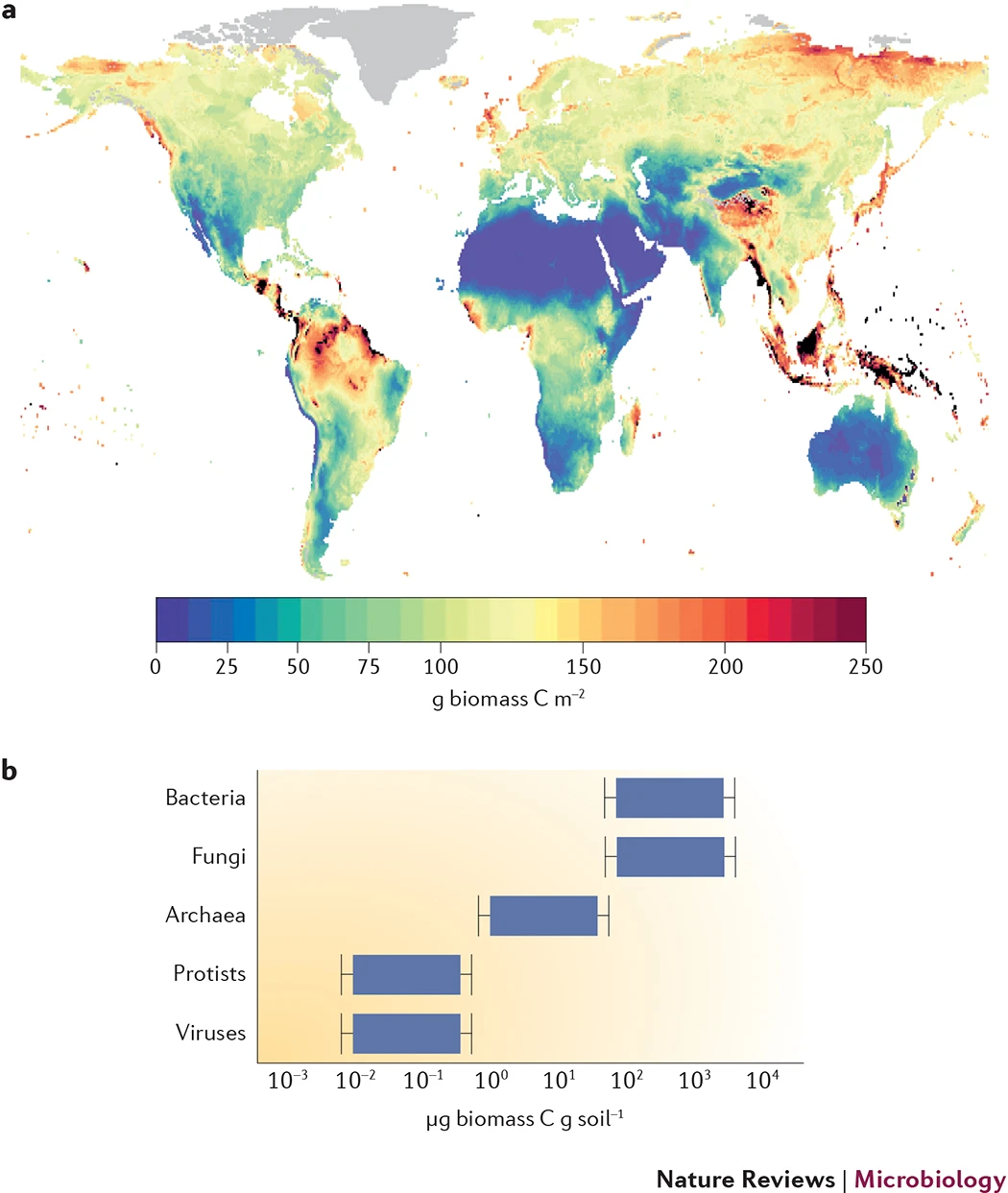
圖3 土壤細(xì)菌、古菌、原生生物和真菌群落結(jié)構(gòu)
Figure 3 The general structure of the bacterial, archaeal, protistan and fungal communities found in soil. The data show the range in proportional abundances for each major group across the 66 unique soil samples described by Crowther et al.40. Abundances were estimated by marker gene sequencing (16S ribosomal RNA (rRNA) for bacteria and archaea, internal transcribed spacer 1 (ITS1) for fungi and the 18S rRNA gene for protists). All soils were collected from relatively undisturbed sites (no cultivated soils) from across North America. The soils represent a wide range of ecosystem types (including both forests and grasslands), latitudes (from 18°N to 65°N), soil characteristics and climatic conditions. MBGA, Marine Benthic Group A.
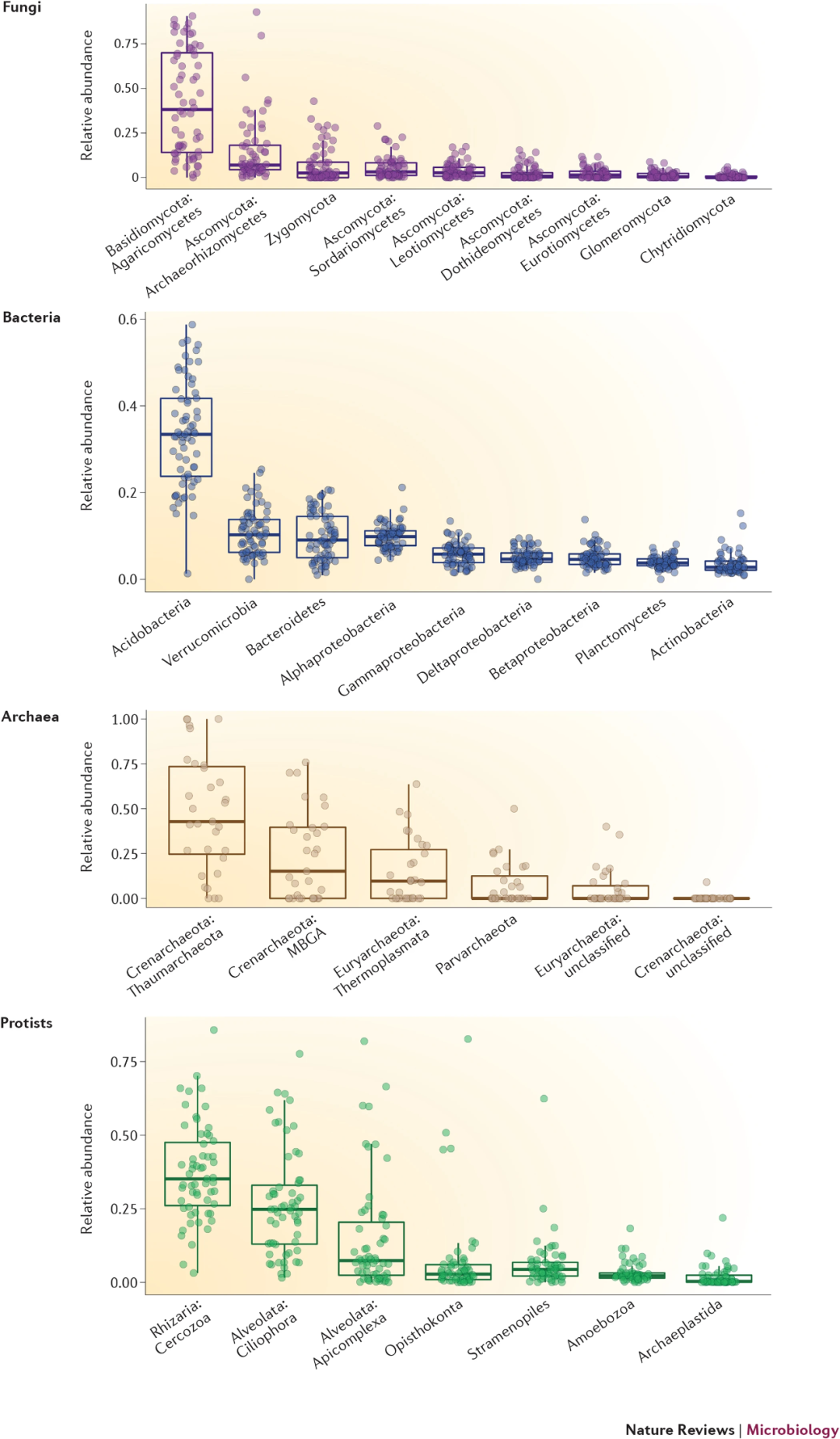
圖4 影響土壤細(xì)菌群落組成的生物和非生物因素
Figure 4 Biotic and abiotic factors that can influence the composition of soil bacterial communities. A hierarchy of biotic and abiotic factors that can influence soil bacterial communities, and their relative importance in influencing the structure of soil bacterial communities across space or time. 'Importance' is defined here as the ease of detecting the effects of these factors on the overall composition of soil bacterial communities. These factors are not necessarily independent and can correlate with one another (for example, soil texture can influence soil moisture availability). Furthermore, the importance of these factors will depend on the soils under investigation and the bacterial lineage in question. The shading of each box qualitatively indicates how well we understand the specific effects of each factor on bacterial communities; darker shades highlight factors that have been reasonably well-studied. This hierarchy is based primarily on studies that have examined spatial patterns in soil microbial communities.
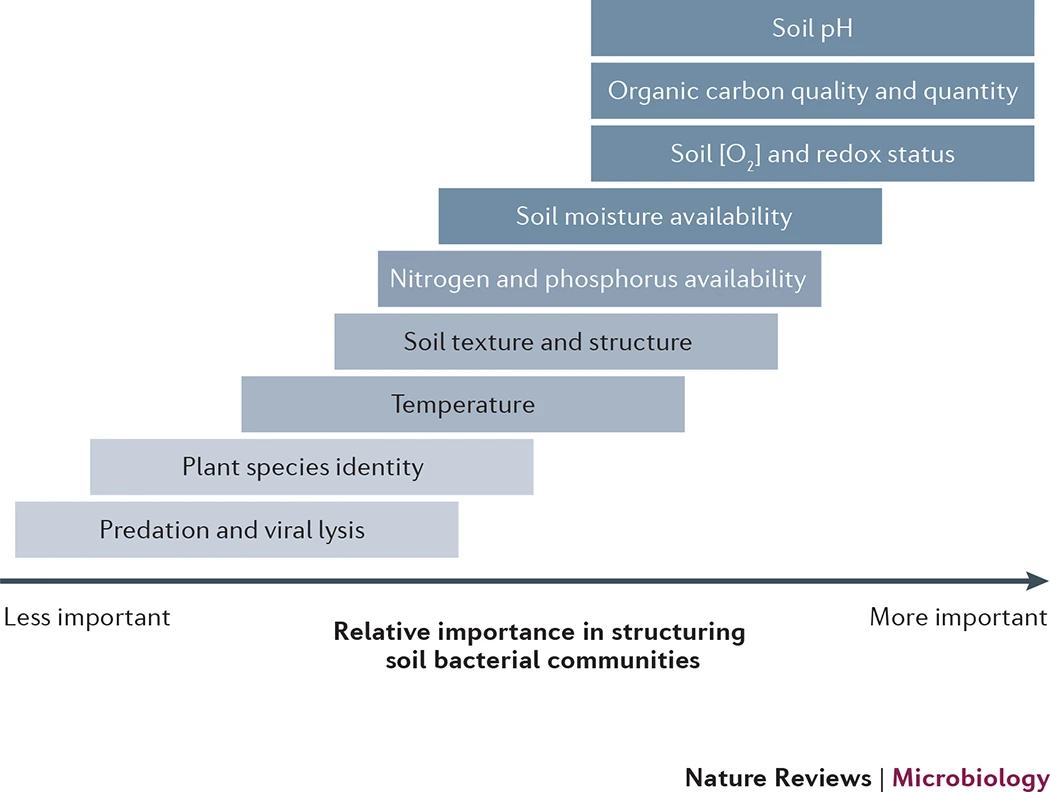
圖5 由土壤微生物組調(diào)控的土壤生物地球化學(xué)過程
Figure 5 Soil biogeochemical processes that can be modulated by the soil microbiome. A summary diagram that highlights a subset of the important soil biogeochemical processes that are directly modulated by soil microorganisms. The vertical arrows indicate microbial processes that are responsible for the production or consumption of trace gases at the soil–atmosphere interface. The curved arrows indicate some of the key microbial processes that can occur within soil, processes that can regulate soil acidity, the availability of nitrogen, phosphorus or other nutrients, and the lability (ease of consumption by microorganisms) of soil organic carbon pools. Non-methane volatile organic compounds (VOCs) include acetone, methanol, formaldehyde, isoprene and other organic compounds with low molecular weight. As indicated in the text, this Figure just highlights a subset of all possible soil microbial processes and does not highlight the inter-related nature of these processes, the specific metabolic pathways responsible, or the range of direct and indirect mechanisms by which soil microbial symbionts and pathogens can influence plants. The shading of the arrows indicates which processes are expected to be carried out by a relatively small subset of microbial taxa (light grey; 'narrow' processes), by an intermediate number of taxa (dark grey) and by a broad diversity of taxa (black; 'broad' processes).
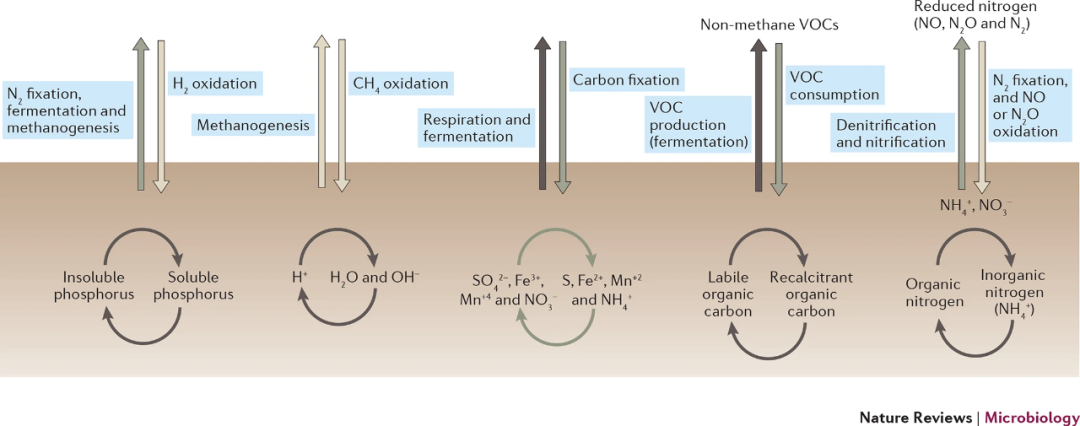
圖6 Grime提出的competitor–stress tolerator–ruderal(CSR)框架應(yīng)用于土壤細(xì)菌異養(yǎng)菌
Figure 6 Grime's competitor–stress tolerator–ruderal framework applied to soil bacterial heterotrophs. An outline of Grime's competitor–stress tolerator–ruderal (CSR) framework and how it may apply to soil bacterial heterotrophs. Boxes include selected phenotypic or genomic attributes that may be associated with the three broad categories of life-history strategies along with examples of soil bacterial groups that are likely to fall into these categories. These categories are not meant to be discrete; rather, they represent a conceptual framework that helps to organize phenotypic or genomic information regarding the adaptive strategies that are used by soil bacterial taxa to survive in soil. μmax, maximum potential growth rate; EPS, extracellular polymeric substances; rRNA, ribosomal RNA.
山東靠山生物科技有限公司轉(zhuǎn)載!

